Make a Scatterplot for Benchmarking
make_benchmarking_scatterplot(result_list, ref_data, file_name = NULL)Arguments
Value
the ggplot object
Examples
data("single_cell_data_1")
data("cell_type_annotations_1")
data("batch_ids_1")
data("bulk")
data("RefData")
common_genes <- intersect(rownames(single_cell_data_1), rownames(bulk))[1:2000]
single_cell_data <- single_cell_data_1[common_genes, 1:500]
cell_type_annotations <- cell_type_annotations_1[1:500]
batch_ids <- batch_ids_1[1:500]
bulk <- bulk[common_genes, ]
RefData <- RefData[, order(colnames(RefData))]
res_bisque <- deconvolute(
bulk, NULL, "bisque", single_cell_data,
cell_type_annotations, batch_ids
)
res_scdc <- deconvolute(bulk, NULL, "scdc", batch_ids,
single_cell_object = single_cell_data,
cell_type_annotations = cell_type_annotations
)
#> Creating Basis Matrix adjusted for maximal variance weight
#> Used 1630 common genes...
#> Used 6 cell types in deconvolution...
#> HD3021b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c0 has common genes 1559 ...
#> WNNLS Converged at iteration 72
#> HD3021b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c1 has common genes 1556 ...
#> WNNLS Converged at iteration 3
#> HD3021b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c3 has common genes 1565 ...
#> WNNLS Converged at iteration 10
#> HD3021b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c7 has common genes 1559 ...
#> WNNLS Converged at iteration 14
#> HD3121b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c0 has common genes 1571 ...
#> WNNLS Converged at iteration 4
#> HD3121b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c1 has common genes 1556 ...
#> WNNLS Converged at iteration 1
#> HD3121b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c3 has common genes 1546 ...
#> WNNLS Converged at iteration 1
#> HD3121b2ee377f8711ec9bf265fb9bf6ab9cPBMC21b2ee377f8711ec9bf265fb9bf6ab9c7 has common genes 1558 ...
#> WNNLS Converged at iteration 3
result_list <- list(SCDC = res_scdc, Bisque = res_bisque)
# Merging the two T cell props
result_list <- lapply(result_list, function(x) {
cbind(x, T = (x[, "CD4 T"] + x[, "CD8 T"]))[, -c(2, 3)]
})
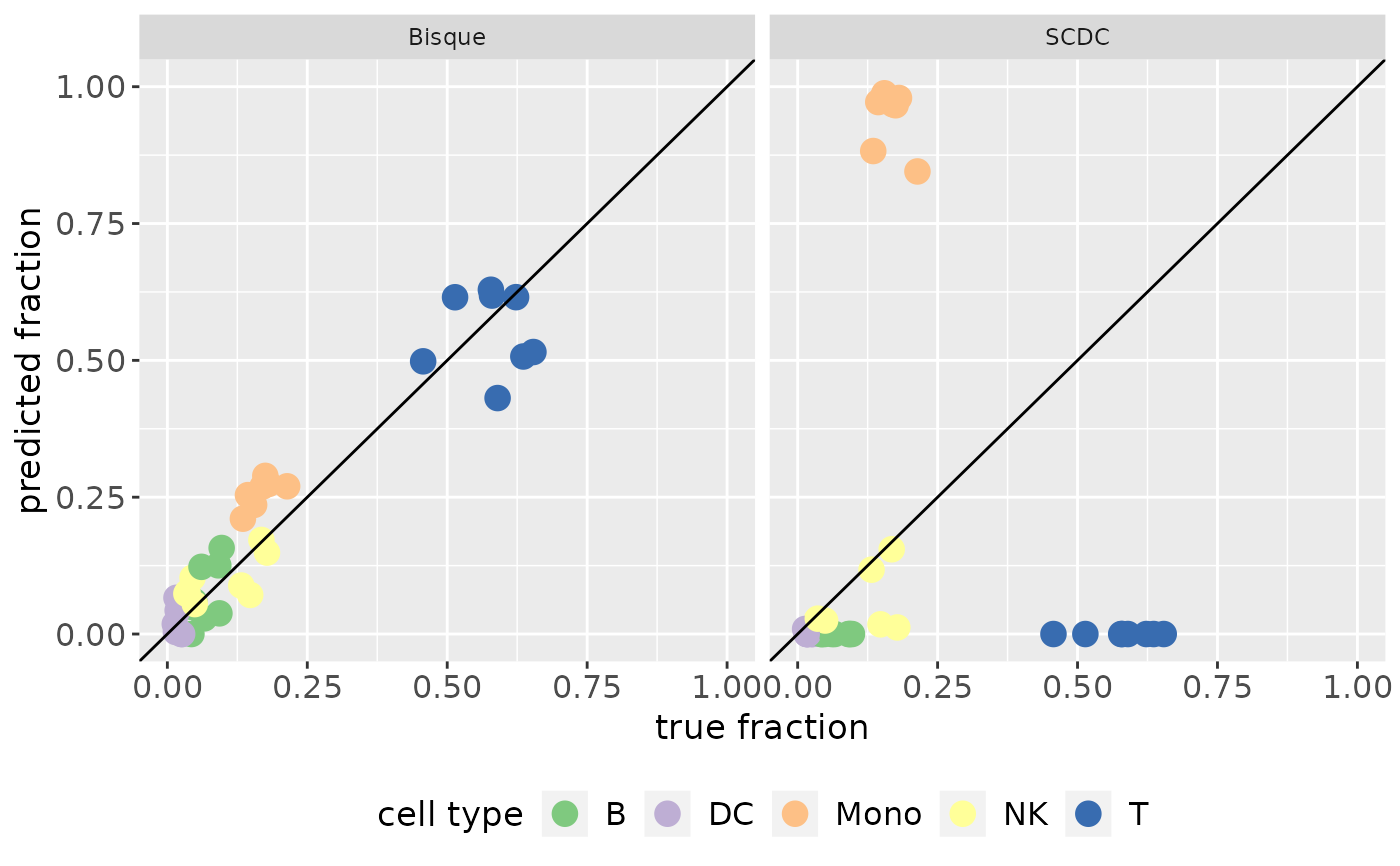
make_benchmarking_scatterplot(result_list, RefData)
 # Alternative if you want to save the plot in a file
# make_benchmarking_scatterplot(result_list, RefData, "predictionVsGroundtruth.png")
# Alternative if you want to save the plot in a file
# make_benchmarking_scatterplot(result_list, RefData, "predictionVsGroundtruth.png")